| [ Team LiB ] |
|
10.1 NCBI-BLAST InstallationNCBI-BLAST, as the name implies, is available from the National Center for Biotechnology Information (NCBI). Precompiled binaries and source code are available for free and without restriction. The source code is in the public domain, so there are quite a few derivative works, both commercial and free (see Chapter 12). NCBI-BLAST is currently available as precompiled binaries for 11 popular operating system-hardware combinations. In addition, there is this very generous statement in the README.bls file:
If you have a platform that isn't supported as a precompiled binary, you may wish to take up the offer from the NCBI, or you may be able to find one using an Internet search engine such as Google. You can also compile the executables yourself; the source code may be obtained as part of the NCBI toolbox: ftp://ftp.ncbi.nih.gov/toolbox/ncbi_tools. For more information about the toolbox, see http://www.ncbi.nlm.nih.gov/IEB/ToolBox. This chapter will take you through the installation procedures for Unix, Windows, and Macintosh. It doesn't cover how to build the NCBI executables from source. If you are a Windows or Macintosh user, please read the Unix installation first because it has some information that isn't duplicated in the other sections. 10.1.1 Unix InstallationThe first step is to download a compressed Unix tape archive, often called a tarball, to your computer. Find the appropriate executable for your system at ftp://ftp.ncbi.nih.gov/blast/executables. A note of caution here: the files in the tarball aren't contained in a subdirectory so it is a good idea to place the tarball in its own directory before you expand the archive. If you're downloading via a browser, you may have plug-ins that automatically expand the archive. This could leave you with a bunch of files all over your system, or it may create a directory for you. To be safe, if you're using a browser, download the tarball to a new directory, for example, /usr/local/pkg/ncbi-blast, or perhaps ncbi-blast in your home directory if you don't have root access. If the archive hasn't already been expanded, you can expand it with this command, where your_platform_name will be something like linux.tar.Z or linux.tar.gz: tar -xzf blast.your_platform_name Not all versions of tar support the -z option above, in which case you can use the following command line: zcat blast.your_blastform_name | tar -xf - 10.1.1.1 Files and directoriesMore than 20 files come with the installation. Table 10-1 shows the files and a very brief description in logical order. See the NCBI-BLAST reference in Chapter 13 for comprehensive coverage of each program.
10.1.1.2 The .ncbirc fileThe next step is to create a resource file that tells blastall where to find its scoring matrices and other related files. The contents of the file are just these two lines: [NCBI] Data="/usr/local/pkg/ncbi-blast/data/" You may also add to this file a line giving the location of the BLAST database files. [BLAST] BLASTDB=path_to_db This file must be named .ncbirc (including the leading dot) and should be located in every user's home directory (although it can also be in the directory where blastall resides). 10.1.1.3 Setting the PATH and BLASTDB environment variablesThe next step is to make sure the programs can be called without explicit paths—that is, without having to type the full pathname every time you want to run the program. You should either place symbolic links from the executables in /usr/local/bin or modify your PATH environment variable. If you're not sure how to do this, ask your Unix system administrator to help you or consult an introductory Unix book. The final step allows you to select databases by name rather than by explicit path. This is more than just a convenience; the abstraction also lets you provide a similar interface on multiple machines where the underlying directory structure may be different. Here is an example of what you might put in your .cshrc file if you use csh or its derivatives as your shell: setenv BLASTDB /usr/local/blastdb If you're using one of the sh derivatives, such as bash, use the following: export BLASTDB=/usr/local/blastdb That's it, except that you can't use the software without sequences. If you don't need to know about Mac or Windows installation, skip ahead to the command-line tutorial. 10.1.2 Windows InstallationDownload the blastz.exe file from ftp://ftp.ncbi.nih.gov/blast/executable, and place this in its own directory, such as C:\ncbi-blast\. This is a self-extracting archive, so you can simply double-click on it, and all the files will be extracted into the current directory. See Table 10-1 for a description of all the files. 10.1.2.1 The ncbi.ini fileSimilar to the Unix install, a special file must be created with the path to the data directory. Create a file called ncbi.ini in either the Windows or WINNT directory with the following contents: [NCBI] Data="C:\ncbi-blast\data" Unlike Unix, rather than setting the BLASTDB environment variable to the location of the BLAST databases, add the following to the ncbi.ini file. [BLAST] BLASTDB="C:\ncbi-blast\db" 10.1.2.2 Setting the PATH environment variableThe PATH environment variable works like its Unix counterpart. The easiest way to set it is to right-click on the My Computer icon, click on the Advanced tab, and then click on the Environment Variables button (Figure 10-1). Figure 10-1. Selecting the PATH environment variable This brings up the System Variables window. Select the Path variable to edit and add ;C:\ncbi-blast to the end of the Path (Figure 10-2) Note that there's a semicolon before the C, which is the separator between directories. Now the BLAST executables can be used from any DOS prompt. Figure 10-2. Adding the PATH environment variable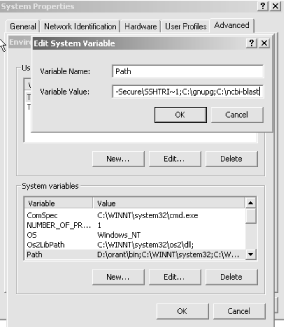 10.1.3 Macintosh OS X InstallationMacOS X is Unix under the hood, so you can follow the previous Unix installation procedures (the file is called blast.darwin.tar.Z because Darwin is the actual name of the Unix that MacOS X uses). Alternatively, you can use the friendly installer available from the folks at http://bioteam.net who have put together a CD containing quite a few common bioinformatics application suites including Apple-Genentech-BLAST (an optimized version of NCBI-BLAST, see Chapter 12). The CD image is located at http://gm.sonsorol.org:8080/BioInfxToolsInstaller.cdr. The installation procedure could not be much simpler. Double-click on the BioInfxToolsInstaller.cdr image, open the BioInfxToolsInstaller that appears on your desktop, and then double-click the agncbi12-20-2001.pkg. This launches a typical installer, and after a few clicks and keystrokes, you're done. At the end, you need to do two more things: add one line to your .cshrc file and copy the .ncbirc file to your home directory. To do this, open the Terminal application and type the following two lines exactly as they appear here: echo "source/usr/local/biotools/cshrc.biotools" >> ~/.cshrc cp /usr/local/biotools/.ncbirc ~/.ncbirc 10.1.4 Macintosh OS 9 InstallationThe OS 9 archive is called blast.hqx. If you click on the file icon, your browser will most likely launch the appropriate tools to automatically expand the archive. If not, you can use Stuffit Expander, which is available for free from http://www.stuffit.com. The OS 9 applications look completely different from the command-line versions because they all have a graphical interface. Don't worry about this because the interface isn't pretty, and you have to drag the window across your screen several times to see all the buttons and text fields. (You may also experience a few system crashes because OS 9 isn't the ideal environment for BLAST.) You must also create a special file to tell BLAST where to find its data directory. Create a file called ncbi.cnf in your system folder that contains the path to the data folder. For example, if the data folder is in a computer named MyMac and in a folder called Blast, the ncbi.cnf file should look like this: [BLAST] BLASTDB=MyMac:Blast:data Installation instructions for OS 9 are included for completeness, but Apple no longer supports this operating system. You might want to upgrade to OS X or install one of the Linux distributions for PPC. If you install Linux, you may have to compile the executables from the source, but it's worth checking if anyone has already done this. A Google search for "Mac linux BLAST" is a good place to start. |
| [ Team LiB ] |
|