| [ Team LiB ] |
|
A.2 Detailed Descriptions and ExamplesThis section includes detailed descriptions of each format, followed by an example. To create the examples, the authors performed a BLASTP search of the coelacanth HoxA11 protein sequence (AAG39070) versus the HoxDB.pep database, which is included in the online supplement. A.2.1 Option 0: Pairwise AlignmentsOption 0 is the default alignment and the classic BLAST format. The definition line of the subject is given at the top of each entry, marked with the greater-than sign (>) and followed with the subject's total length. For each HSP of a subject, the score, expect, identities, positives, and gaps are reported and followed by a pairwise alignment. For the pairwise alignment in Figure A-1, the query sequence is shown on the first row and the subject on the third row. Gaps are represented in each as a dash (-). Between the query and subject lies the alignment row, which shows the residue for identities, a plus (+) for positive scoring alignments, and a dot (.) for mismatches. In BLASTN alignments, the middle row has vertical bars (|) for identities and nothing for mismatches. Figure A-1. Option 0: Standard pairwise alignment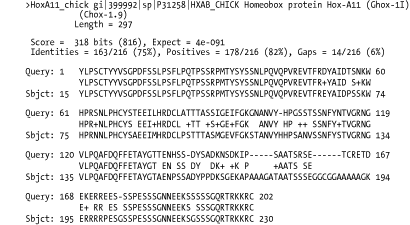 A.2.2 Query-Anchored AlignmentsAll query-anchored formats (1-6) are multiple-sequence alignments. They share the same general form, with the query repeated at the top of each line and all matching subjects aligned on subsequent lines. The difference between showing identities and not showing them is counterintuitive. For the options that show identities (1 and 3), identical residues are symbolized with a dot (.), similar amino acids are in uppercase, and mismatches are in lowercase. For the options without identities (2, 4, 5 and 6) every residue is shown with identities and similar residues in uppercase and mismatches are in lowercase. A.2.3 Option 1: Query-Anchored Showing IdentitiesIn the format shown in Figure A-2, the identical residues are represented by a dot (.) and insertions and deletions are represented in the subject sequences, but not the query. Figure A-2. Option 1: Query-anchored showing identities A.2.4 Option 2: Query-Anchored, No IdentitiesThis format (Figure A-3) is the same as Option 1 (Figure A-2), but all residues are shown with identities and positives in uppercase and mismatches in lowercase. As with Option 1, insertions and deletions are represented in the subject sequences, but not the query. Figure A-3. Option 2: Query-anchored, no identities A.2.5 Option 3: Flat Query-Anchored Showing IdentitiesSame as Option 1 (Figure A-2), but insertions or deletions in Figure A-4 are padded in the query, rather than shown in the subjects. This is a more compact format than the nonflat one, which has residues dangling down to represent insertions within the subject sequences. Figure A-4. Option 3: Flat query-anchored showing identities A.2.6 Option 4: Flat Query-Anchored, No IdentitiesThis format is the same as Option 2 (Figure A-3), but insertions or deletions in Figure A-5 are padded in the query, rather than shown in the subjects. Thus, the entire multiple sequence alignment is flat, without subject insertions dangling down. Figure A-5. Option 4: Flat query-anchored, no identities A.2.7 Option 5: Query-Anchored, No Identities, and Blunt EndsBlunt-end options extend the HSPs out to the beginning and end of the entire query sequence so that each HSP is shown in all lines of the alignment. In Figure A-6, the HoxD11_chick and HoxD11_human entries have additional HSPs that are seen later in the alignment (not shown). You see the dashes (-) at the beginning of the second HSP of each, which makes the entry blunt. Figure A-6. Option 5: Query-anchored, no identities and blunt ends A.2.8 Option 6: Flat Query-Anchored, No Identities, and Blunt EndsSame as Option 5 (Figure A-6), but the insertion and deletion characters in Figure A-7 are inserted into the query, making it flat, without any dangling insertions in the subject alignment lines. Figure A-7. Option 6: Flat query-anchored, no identities and blunt ends A.2.9 Option 7: XMLThe BLAST eXtensible Markup Language (XML) is specified by the Data Type Definition (DTD) file, NCBI_BlastOutput.dtd, which is located at http://www.ncbi.nlm.nih.gov/dtd/. This format isn't meant to be human-readable, but Figure A-8 shows the first few lines from the BLASTP search so you can get a feel for how XML looks. Figure A-8. Option 7: XML format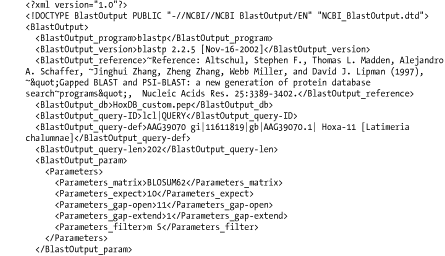 A.2.10 Option 8: Tabular, Without Comment LinesTabular formats are very nice for easy parsing. All fields in Figure A-9 are tab-delimited. The fields are query id, subject id, percent identity, alignment length, mismatches, gap openings, query start, query end, subject start, subject end, e-value, and bit score. Figure A-9. Option 8: Tabular, without comment lines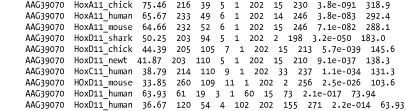 A.2.11 Option 9: Tabular, with Comment LinesThe format of Figure A-10 is the same as that in Option 8 (Figure A-9), except it includes four header lines at the top of each query that describe the BLAST program, the query, the database, and the fields of the alignment. Figure A-10. Option 9: Tabular, with comment lines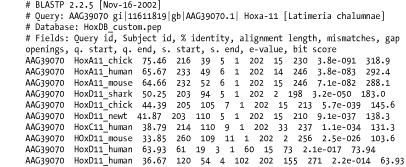 A.2.12 Option 10: ASN.1 Text FormatAbstract Syntax Notation One (ASN.1) is an International Standards Organization (ISO) data format. ASN.1 is used to mark up data for reliable, robust exchange. Like XML, it isn't meant to be human-readable, but Example A-1 shows the first few lines of a BLASTP search so you can get a feel for the syntax. For more information on the NCBI use of ASN.1, see http://www.ncbi.nlm.nih.gov/Sitemap/Summary/asn1.html. Example A-1. ASN.1 text formatSeq-annot ::= {
desc {
user {
type
str "Hist Seqalign" ,
data {
{
label
str "Hist Seqalign" ,
data
bool TRUE } } } ,
user {
type
str "Blast Type" ,
data {
{
label
str "BLASTP" ,
data
int 2 } } } } ,
data
align {
{
type partial ,
dim 2 ,
score {
{
id
str "score" ,
value
int 699 } ,
{
id
str "e_value" ,
value
real { 139321249, 10, -85 } } ,
{
id
str "bit_score" ,
value
real { 273862735, 10, -6 } } ,
{
id
str "num_ident" ,
value
int 140 } } ,
A.2.13 Option 11: ASN.1 Binary FormatThis option produces the same ASN.1 output as Option 10, but in binary format. It isn't readable and therefore isn't shown. |
| [ Team LiB ] |
|