13.8 blastpgp Parameters (PSI-BLAST and PHI-BLAST)
blastpgp
is the program used to run PSI-BLAST and PHI-BLAST. These programs
are specialized protein BLAST comparisons that are more sensitive
than the standard BLASTP search. PSI-BLAST considers
position-specific information when searching for significant hits.
PHI-BLAST uses a pattern, or profile, to seed an alignment, which is
then extended by the normal BLASTP algorithm.
13.8.1 PSI-BLAST
PSI-BLAST
(position-specific iterated BLAST) uses a specialized scoring matrix
that assigns scores to each position (hence, position-specific) in
the query sequence based on alignments defined by consecutive
iterations of searches (hence, iterated). The specialized matrix is a
position-specific scoring matrix (PSSM) that assigns a score for
every amino acid at each position in the query sequence (See Figure 13-1).
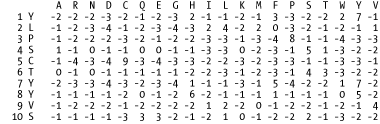
Figure 13-1 shows a portion of a PSSM calculated for
the coelacanth Hoxa11 protein (AAG39070). The query amino acids are
numbered in the left column with the position-specific scores for
each of the 20 amino acids shown across each row. The diverse scores
of the three Tyrosines (Y) at positions 1, 7, and 8 highlight the
position-specific aspect of this scoring scheme compared to
traditional BLAST matrices, which would contain the same scores for Y
in all three positions.
The PSSM, or checkpoint file, is
created internally by PSI-BLAST, but it can also be exported to a
file using the -C option of
blastpgp. This option is extremely useful. You
can use the checkpoint file in subsequent PSI-BLAST
(blastpgp) searches or as a database entry for
the RPS-BLAST program. You can also use the PSSM in a specialized
tblastn search in blastall
by using the -p psitblastn and -R
<checkpoint file> options with a nucleotide database.
To run PSI-BLAST, the
-j parameter must be set to something greater than
1. The default of -j 1 means
that there are no iterations and that it's therefore
the same as a single BLASTP search. Setting -j
sets the maximum number of iterations to run, with the program
stopping beforehand if the search comes to convergence. Convergence
occurs when no new sequences are found that are better than the E
value threshold set by the -h parameter.
Here are a few sample command lines:
blastpgp -d nr -i my_protein -s T -j 5
blastpgp -d nr -i my_protein -R my_protein.ckp -d nr -j 5 -h 0.001
13.8.2 PHI-BLAST
PHI-BLAST stands for pattern-hit
initiated BLAST. The program uses an input sequence and a defined
pattern to query a protein database. The pattern is defined in
PROSITE format (http://ca.expasy.org/prosite/)and is used as the seed for the alignment. The pattern
is used instead of the words that are usually generated for seeding
alignments in BLASTP. Here's a sample profile:
ID HoxA11 pattern1
PA Y-S-[SA]-X-[LVIM]
The profile's syntax has
a line starting with ID, followed by two spaces
and the name of the pattern. The name is free text. The next line
should start with PA, followed by two spaces, and
then the pattern in PROSITE format. The PROSITE format is simple. A
dash (-) separates letters, an X means any letter, and the brackets
([]) specify a choice of amino acids. You can find more information
on the pattern syntax in the README.bls file
that comes with the NCBI-BLAST distribution.
Additionally, if the pattern occurs
more than once in the query and you would like to limit which
occurrences are used as seeds, specify those locations by using the
HI (hit initiation) tag in the pattern file. You set
-p to seedp instead of
patseedp (explained in the reference section
that follows). The following example specifies that the pattern
starting at position 143 should be used. (In this case,
there's also an occurrence at 34, which is ignored.)
ID HoxA11 pattern2
PA Y-S-[SA]-X -[LVIMK]
HI 143
PHI-BLAST can also be a jumping-off point for a PSI-BLAST run. In the
following command line, the pattern in hit_file
initiates the first iteration of PSI-BLAST for the development of the
PSSM, followed by normal rounds of PSI-BLAST iterations.
blastpgp -d nr -i my_protein -k hit_file -p patseedp -j 5
Here are a few sample PHI-BLAST command lines:
blastpgp -d nr -i my_protein -k hit_file -p patseedp
blastpgp -d nr -i my_protein -k multi_hit_file -p seedp
blastpgp -d HoxDB.pep -i AAG39070.pep -k hit_file.hox -p patseedp
The following reference describes parameters used with
blastpgp, which executes PSI- and PHI-BLAST
searches.
The number of processors to use; same as
blastall.
| Default: blastn 0, others 40 | |
The multiple-hit window size; same as blastall.
The number of alignments to show;
same as blastall.
| Default: Optional | Program: PSI-BLAST only |
The input alignment file for a PSI-BLAST restart. It allows a
PSI-BLAST run to start with a curated multiple sequence alignment
instead of allowing the program to generate it from the first round
of database alignments. For example:
blastpgp -i query -B multiple_alignment -j 5 -d nr
The
alignment file must be based on the Clustal format but without the
header and footer. The file should have a row for each sequence and
can be broken into blocks separated by one or more blank lines. The
query file (specified by -i) must be included in
the alignment (though it doesn't need to be the
first one), and all rows must be padded with dashes (—-) to
make them equal lengths. Also, each column must contain either all
uppercase or lowercase letters. An uppercase letter signifies that
the column should be given a position-specific score; a lowercase
letter means that the matrix (specified by -M)
score should be used. Here is a portion of the example alignment file
included in README.bls (the query is 26SPS9_Hs, in this case):
26SPS9_Hs IHAAEEKDWKTAYSYFYEAFEGYdsidspkaitslkymllc
F57B9_Ce LHAADEKDFKTAFSYFYEAFEGYdsvdekvsaltalkymll
YDL097c_Sc ILHCEDKDYKTAFSYFFESFESYhnltthnsyekacqvlky
YMJ5_Ce LYSAEERDYKTSFSYFYEAFEGFasigdkinatsalkymil
FUS6_ARATH KNYIRTRDYCTTTKHIIHMCMNAilvsiemgqfthvtsyvn
COS41.8_Ci SLDYKLKTYLTIARLYLEDEDPVqaemyinrasllqnetad
644879 KCYSRARDYCTSAKHVINMCLNVikvsvylqnwshvlsyvs
YPR108w_Sc IHCLAVRNFKEAAKLLVDSLATFtsieltsyesiatyasvt
eif-3p110_Hs SKAMKMGDWKTCHSFIINEKMNGkvw---------------
T23D8.4_Ce SKAMLNGDWKKCQDYIVNDKMNQkvw---------------
YD95_Sp IYLMSIRNFSGAADLLLDCMSTFsstellpyydvvryavis
KIAA0107_Hs LYCVAIRDFKQAAELFLDTVSTFtsyelmdyktfvtytvyv
F49C12.8_Hs LYRMSVRDFAGAADLFLEAVPTFgsyelmtyenlilytvit
Int-6_Mm KFQYECGNYSGAAEYLYFFRVLVpatdrnalsslwgklase
26SPS9_Hs kimlntpedvqalvsgklalryagrqtealkcvaqasknr
F57B9_Ce ckvmldlpdevnsllsaklalkyngsdldamkaiaaaaqk
YDL097c_Sc mllskimlnliddvknilnakytketyqsrgidamkavae
YMJ5_Ce ckimlneteqlagllaakeivayqkspriiairsmadafr
FUS6_ARATH kaeqnpetlepmvnaklrcasglahlelkkyklaarkfld
COS41.8_Ci eqlqihykvcyarvldyrrkfleaaqrynelsyksaihet
644879 kaestpeiaeqrgerdsqtqailtklkcaaglaelaarky
YPR108w_Sc glftlertdlkskvidspellslisttaalqsissltisl
eif-3p110_Hs ----------------------------------------
T23D8.4_Ce ----------------------------------------
YD95_Sp gaisldrvdvktkivdspevlavlpqnesmssleacinsl
KIAA0107_Hs smialerpdlrekvikgaeilevlhslpavrqylfslyec
F49C12.8_Hs ttfaldrpdlrtkvircnevqeqltggglngtlipvreyl
Int-6_Mm ilmqnwdaamedltrlketidnnsvssplqslqqrtwlih
| Default: 9 | Program: PSI-BLAST only |
Sets a constant in pseudocounts for
PSSM. It's generally not necessary to change this
parameter.
| Default: Optional | Program: PSI-BLAST only |
Outputs a file for PSI-BLAST
checkpointing. This outputs the final PSSM for a multipass run of
PSI-BLAST. The checkpoint file can then be used in a PSI-BLAST
restart (see -R), in a
blastall -p
psitblastn run (also see -R), or as an entry in
an RPS-BLAST database.
blastpgp -d nr -i my_protein -j 5 -C my_protein.ckp
The database name; same as blastall.
The expectation value; same as blastall.
| Default: blastn 2, others 1 | |
The penalty to extend a gap; same as
blastall.
The threshold for extending a hit; same as
blastall.
Filters the query sequence; same as
blastall.
Performs gapped alignment; same as
blastall.
PHI-BLAST requires gapping and therefore forbids -g
F.
| Defaults: blastn 5, others 11 | |
The penalty to open a gap; same as
blastall.
| Default: 0.005 | Program: PSI-BLAST only |
The E-value threshold for inclusion
in PSSM. All alignments better than this threshold are used in
constructing the PSSM.
The end of the required region in query. The default of -1 indicates
the actual end of the query. This option can be used in combination
with -S to specify a particular region to use
The query file; same as blastall.
Shows GIs in defline; same as blastall
The maximum number of passes to use in a multipass version. The
default of 1 is just a regular BLASTP search.
Believes the query definition line; same as
blastall.
| Default: hit_file | Program: PHI-BLAST only |
Specifies the
file containing the PROSITE pattern to be used for seeding in a
PHI-BLAST run. If -k isn't
specified when running PHI-BLAST (e.g. -p
patseedp or -p
seedp), the program looks for a file called
hit_file.
The number of best hits from a
region to keep; same as blastall.
Restricts the search of the database to a list of GIs; same as
blastall.
The cost to decline an alignment.
Alignment view options; same as blastall.
The
matrix; same as blastall.
The number of bits required to trigger gapping.
The output file for alignment; same as blastall.
A SeqAlign file output; same as
blastall.
Specifies whether to run in PSI- or PHI-BLAST mode.
Options
- blastpgp
-
PSI-BLAST mode
- patseedp
-
PHI-BLAST mode. Uses all occurrences
of the hit_file pattern to seed alignments. Any
HI tags (see later) in the
hit_file are ignored.
- seedp
-
PHI-BLAST mode. The specified
pattern is found more than once in the query, and the
hit_file specifies which to use as seeds. The
specific pattern(s) occurrences to use is specified with the
HI tag in the hit_file. For
example, the following hit_file designates
seeding from a pattern that occurs at position 143 of the coelacanth
HoxA11 protein:
ID HoxA11 pattern2
PA Y-S-[SA]-X-[LVIMK]
HI 143
seedp throws an exception if the
hit_file doesn't contain the
HI tags.
Output file for
a PSI-BLAST matrix in ASCII format. This [file]
can't be used in any subsequent programs. Use
-c to output a matrix for subsequent searches.
Input checkpoint file for PSI-BLAST
restart. Uses the checkpoint file. Output with -c.
Calculates locally optimal
Smith-Waterman alignments. Because of the heuristic nature of BLAST,
it sometimes produces nonoptimal local alignments. This option causes
BLAST to run the full Smith-Waterman alignment algorithm on subjects
found by the normal BLAST heuristic. There may be some speed cost
using this option, but it helps guarantee high-quality alignments,
which are important in PSSM generation. Setting -s
T is highly recommended.
The start of the required region in query. Used in combination with
-H, this sets a specific region of the query to be
used when generating the PSSM.
Uses composition-based statistics. With this set to
T, the score is adjusted based on composition
biases in the query and subject sequences. Using it helps avoid
possible corruption of the PSSM because it introduces low-entropy
false positives in the multiple sequence alignment.
Produces HTML output; same as blastall.
Uses
lowercase filtering of a query sequence; same as
blastall.
The number of one-line descriptions to show; same as
blastall.
The word size; same as
blastall.
The X dropoff for gapped alignments; same as
blastall.
X dropoff for ungapped extensions; same as
blastall
The effective length of the search
space; same as blastall.
The effective database size; same as blastall.
The X dropoff for final gapped
alignment; same as blastall.
| 