8.14 Consider Using Ungapped Alignment for BLASTX, TBLASTN, and TBLASTX
The
first versions of BLAST produced strictly ungapped alignments but
were still very useful. Although gapped alignment has some
advantages, it may produce surprising results. When running the
translating BLAST programs (BLASTX, TBLASTN, and TBLASTX), you
generally look for protein coding regions and therefore
don't expect to see stop codons. Stop codons are
very frequent in alignments from these programs, and it
isn't possible to eliminate stop codons by simply
making their scores highly negative. In standard alignment algorithms
(see Chapter 3), no match score can be more negative than the cost of
two gaps. In Figure 8-6, all stop codon scores are
given a value of -999 (for more details, see Chapter 10). Notice how
two alternating gaps skip over the stops in this TBLASTX alignment
between two noncoding sequences (this is a WU-BLAST alignment;
NCBI-BLAST is always ungapped for TBLASTX). You can avoid stop codons
only by using ungapped alignment in addition to highly negative stop
scores. Doing so segments the alignment in Figure 8-6 into three short alignments with insignificant
E-values.

Figure 8-7
demonstrates another feature of gapped alignment: alignments may
extend far beyond the end of an exon because gapped extension is
generally less specific. This is especially annoying in genomes with
short introns in which gapped alignments can extend between
nonadjacent exons and obscure intervening introns and exons. To
reduce these lengthy extensions, decrease X,
increase the gap extension cost, select a more stringent scoring
matrix, or use ungapped alignment.
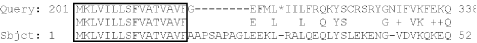
|
