8.7 Know When to Use Complexity Filters
Low-complexity
sequence occurs much more frequently than expected by chance in both
proteins and nucleic acids. When a BLAST search takes longer than
expected, it is almost always due to low complexity sequence or
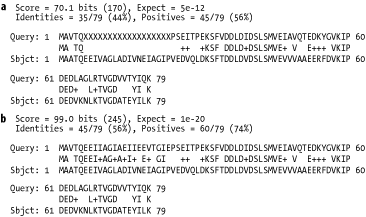
repeats. Low-complexity filters can sometimes be destructive. Figure 8-3a shows what happens when a query sequence is
filtered: the low complexity region is replaced with Xs (or Ns for
nucleotide sequences). This operation always reduces the score and
can terminate an alignment extension. For this reason, it is almost
always better to use soft-masking (see Figure 8-3b). This technique masks low-complexity
sequence in the seeding phase but allows the extension phase to see
the sequence normally. See -F in Chapter 13 and wordmask in Chapter 14.
What if your query is almost
entirely low-complexity? If soft-masking doesn't
work, you may have to perform the search without complexity filters.
In this case, expect many false-positive alignments and a slow
search. Setting a lower E-value to remove low-scoring alignments can
help reduce the size of the output.
 | 